EndoSim is designed in a modular fashion. Input objects
are, in general, lists of functions, where each function accepts a
single numeric vector input ( e.g., temperature, days after
emergence) and returns a numeric vector output of the same length.
For example, in the pest() object, the argument
fun_dev_apt accepts a function describing
temperature-dependent development of nymphs destined to become apterae.
This function should accept as its input daily temperature and return a
corresponding number, ranging from 0-1. The input can be a vector of
length 1:
GPA@fun_dev_apt(20)
#> [1] 0.1376906Or a longer vector:
GPA@fun_dev_apt(c(0, 15, 20, 38))
#> [1] 0.00000000 0.09255006 0.13769057 0.00000000For the supplied pest() object for Myzus
persicae, GPA, this function is currently defined
using the Wang model for development (set up using the internal function
EndoSim:::fit_wang()). However, we can easily replace this
with a different function. This can be done using one of the internal
functions provided with the EndoSim package. For example,
say we want to use a Gaussian function:
GPA@fun_dev_apt <- EndoSim:::fit_gaussian(1, 25, 3)
plot(seq(0, 50, by = 0.1), GPA@fun_dev_apt(seq(0, 50, by = 0.1)), type = "l")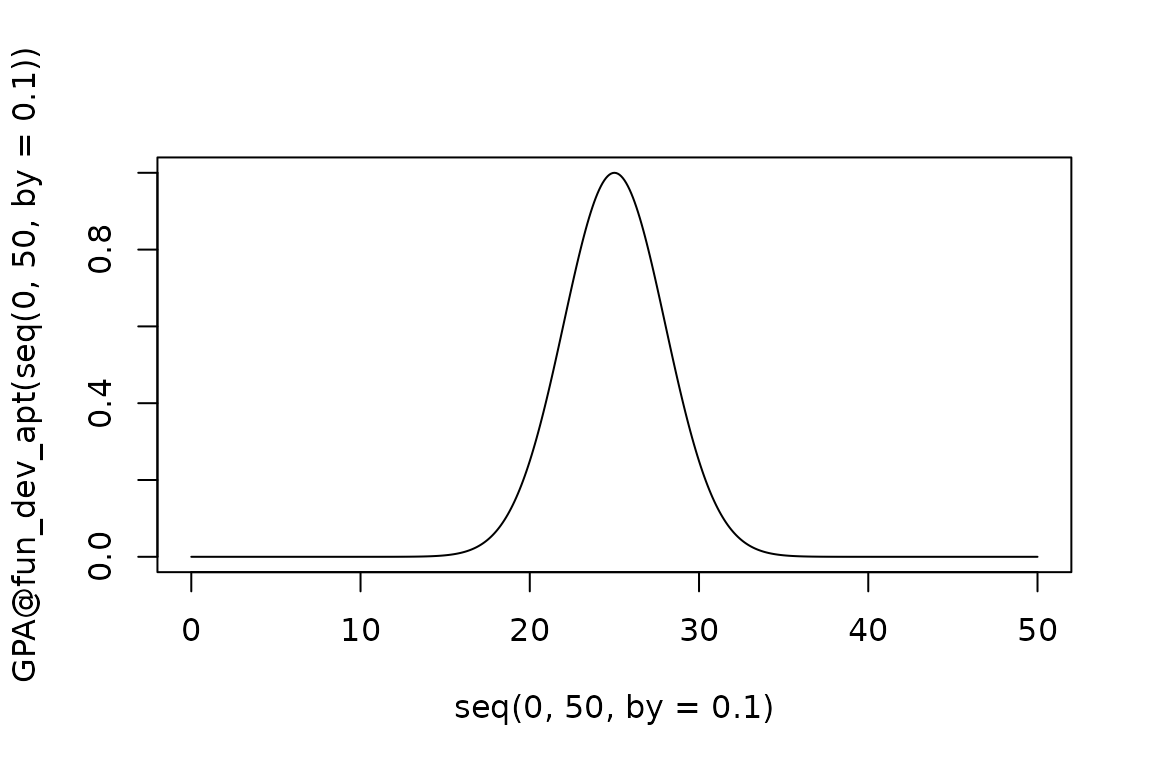
The internal functions come with documentation explaining what the input arguments for each are (generally parameters defining the shape and range of the function), and what the resultant function is - i.e., what is its required input and what is the range of output values it produces. This can make it easy to set up new objects that have similar functional responses as the ones supplied with the package, but may differ in the shape or range. For example, if we want to model a different aphid pest that has narrower thermal preferences and a lower optimal temperature:
GPA@fun_dev_apt <- EndoSim:::fit_gaussian(1, 20, 2)
plot(seq(0, 50, by = 0.1), GPA@fun_dev_apt(seq(0, 50, by = 0.1)), type = "l")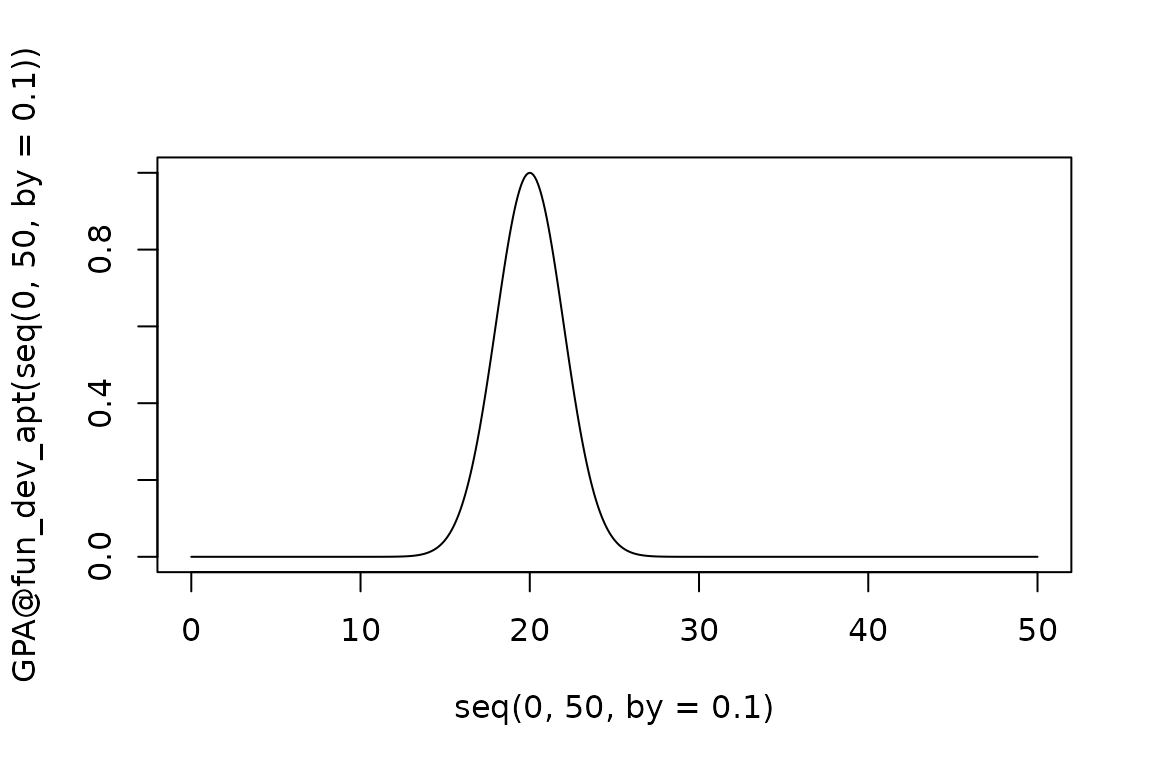
Using the internal functions is not a requirement - we can also write custom functions to represent different processes. However, we strongly urge users to take caution when writing their own functions, since they could potentially return values which are nonsensical or “break” the model. For example, we could write a function where the returned development units change linearly with temperature, but not include bounds which prevent the returned values from being negative:
GPA@fun_dev_apt <- function(temp){
output <- 0.2 * temp - 2
return(output)
}
plot(seq(0, 50, by = 0.1), GPA@fun_dev_apt(seq(0, 50, by = 0.1)), type = "l")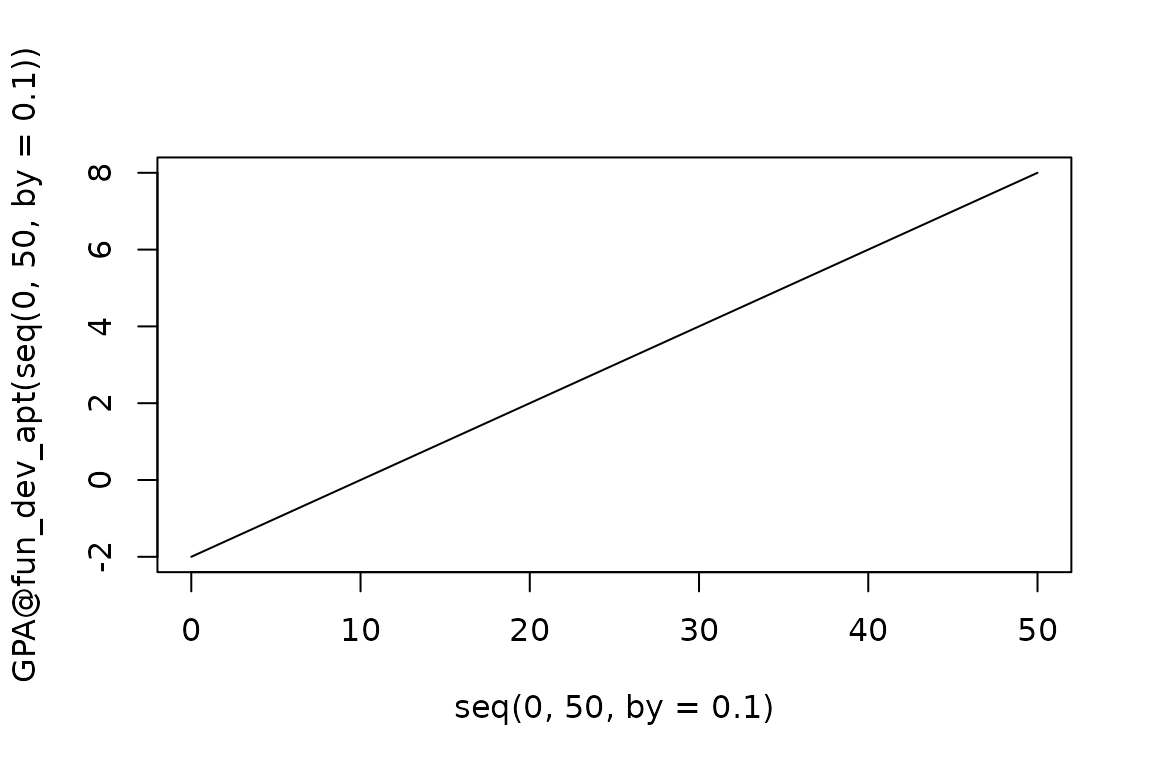
Running a model with this function can lead to downright weird results because of nonsensical development rates:
model <- endosim(Pest = GPA,
Endosymbiont = Rickettsiella,
Crop = Canola,
Parasitoid = DR,
init = init,
conds = Aroonda,
plot = FALSE,
progress = FALSE,
imi = FALSE,
emi = FALSE,
vert_trans = TRUE,
hori_trans = TRUE,
para = TRUE)
plot(model, type = "demo")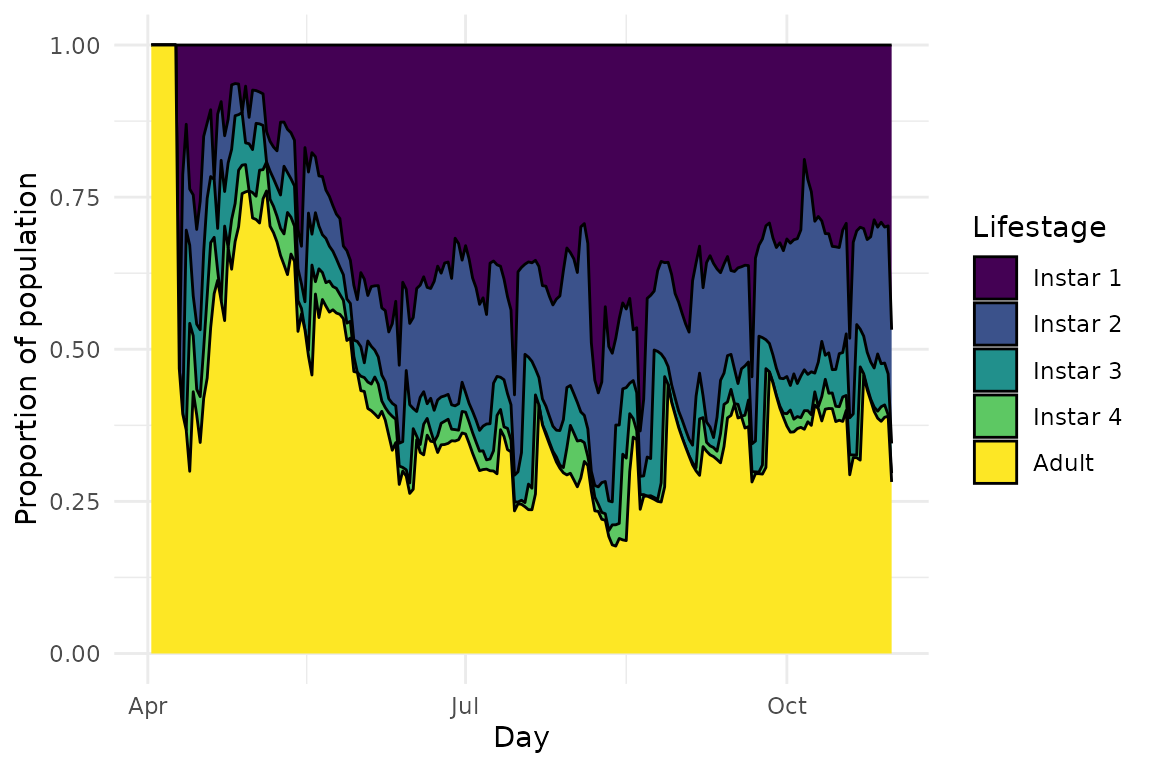
Also note that we could, theoretically, use a function that returns a non-numeric output, or an output of different length to the input. This could simply result in the model failing to run:
GPA@fun_dev_apt <- function(temp){
output <- "this is a text output"
return(output)
}
model <- endosim(Pest = GPA,
Endosymbiont = Rickettsiella,
Crop = Canola,
Parasitoid = DR,
init = init,
conds = Aroonda,
plot = TRUE,
progress = FALSE,
imi = FALSE,
emi = FALSE,
vert_trans = TRUE,
hori_trans = TRUE,
para = TRUE)
#> Error in dev_cohorts[, c(1, 2)] + dev_apt: non-numeric argument to binary operatorTurning processes on and off
Because all objects include functions as arguments, a process can be
easily turned off or fixed ( i.e., made not dependent on its
input) by using the internal EndoSim:::fit_null() function.
This function accepts as an input a user-determined value, and returns a
function which returns this value as an output regardless of the input -
but crucially, of the same length. Some examples:
my_func <- EndoSim:::fit_null(5)
my_func(10)
#> [1] 5
my_func(c(0, 5, 10, 20))
#> [1] 5 5 5 5
my_func("test")
#> [1] 5
my_func <- EndoSim:::fit_null("null")
my_func(10)
#> [1] "null"
my_func(c(0, 5, 10, 20))
#> [1] "null" "null" "null" "null"Because the output is always of the same length, using
fit_null() doesn’t break the model due to inconsistent
lengths of objects.
Turning off a process is as simple as passing to it
fit_null(0) or fit_null(1) - this makes sure
the output for that process is always 0 or 1. Which one to choose
depends on the process, and whether “turning it off” means the returned
value should be 0 or 1.
For instance, disabling age-dependent mortality can be done by
replacing the fun_sen_loss() function so that the returned
value is 0. In this way, there is no mortality associated with the age
of the cohort (senescense), since the number of aphid deaths due to
senescense are always 0, and mortality is modelled as an additive
process.
GPA@fun_sen_loss <- EndoSim:::fit_null(0)
model <- endosim(Pest = GPA,
Endosymbiont = Rickettsiella,
Crop = Canola,
Parasitoid = DR,
init = init,
conds = Aroonda,
plot = TRUE,
progress = FALSE,
imi = FALSE,
emi = FALSE,
vert_trans = TRUE,
hori_trans = TRUE,
para = TRUE)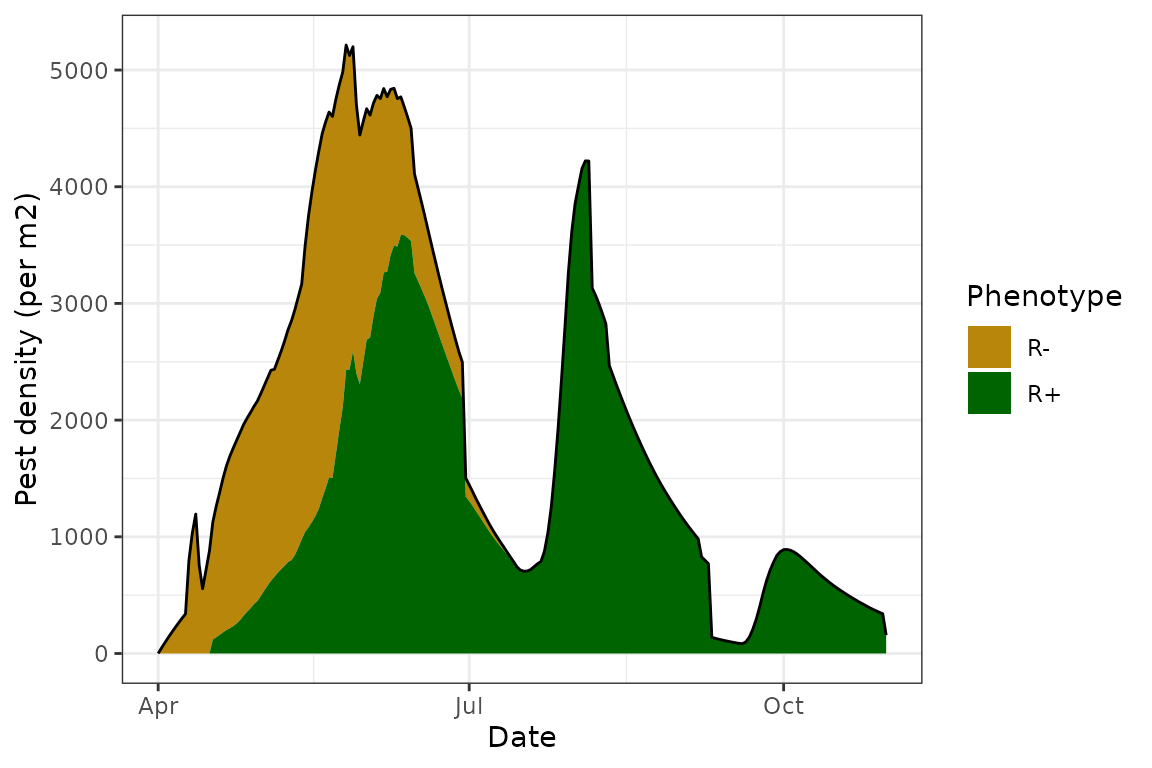
On the other hand, disabling age-dependent reproduction by replacing
the fun_age_fecund() function, requires a value of 1.
Because the total growth rate is a multiplication of temperature-,
density- and age-related contributions, fixing this value to 1
effectively removes the “contribution” of age - growth rates no longer
depend on the age of the cohort.
GPA@fun_age_fecund <- EndoSim:::fit_null(1)
model <- endosim(Pest = GPA,
Endosymbiont = Rickettsiella,
Crop = Canola,
Parasitoid = DR,
init = init,
conds = Aroonda,
plot = TRUE,
progress = FALSE,
imi = FALSE,
emi = FALSE,
vert_trans = TRUE,
hori_trans = TRUE,
para = TRUE)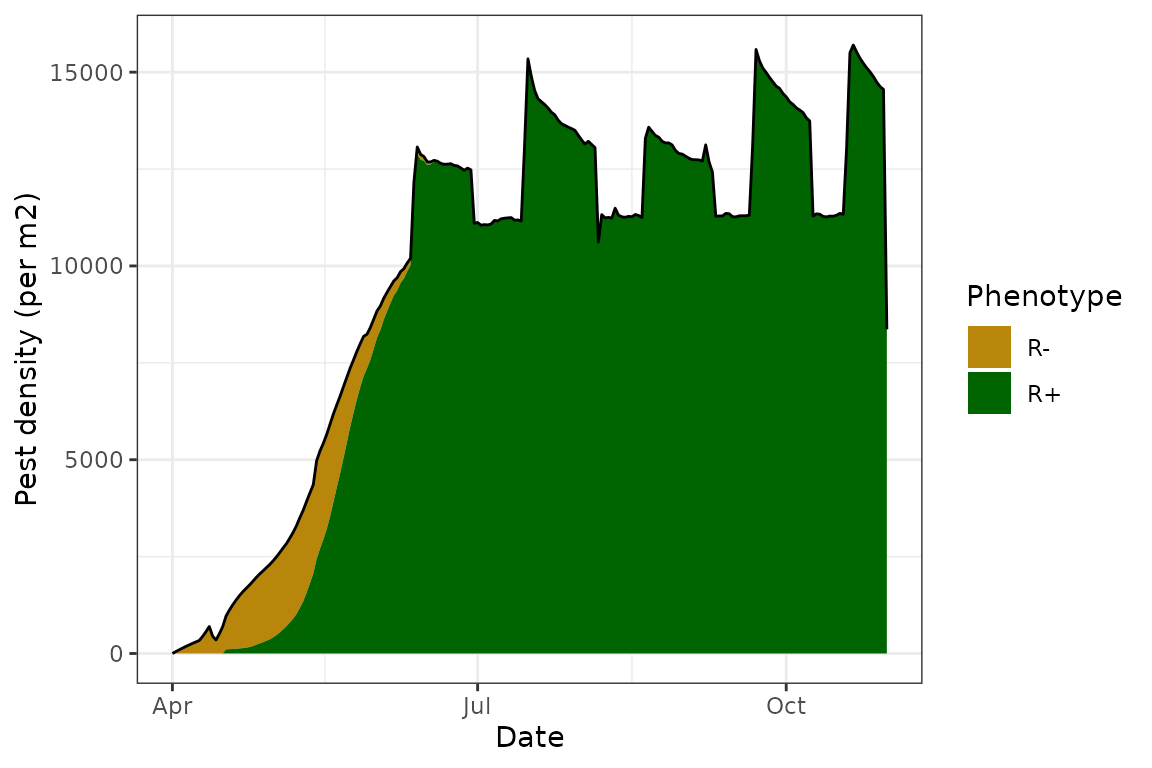
Again - this functionality should be used with caution since it could lead to nonsensical results.
